
| Home About us Mathematical Epidemiology Rweb EPITools Statistics Notes Web Design Search Contact us |
A q-quantile is defined to be the smallest x such that the chance of a random variable being greater than or equal to x is at least q.
The meaning of this is easier to see in the concrete. Let's look back at the binomial again. Let's first plot the cumulative distribution function for the needle example, where the number of trials was 51 and the success probability 0.03:
> plot(plot(0:10,pbinom(0:10,size=51,prob=0.03),ylim=c(0,1)) > abline(h=0.25) > pbinom(0:10,size=51,prob=0.03) [1] 0.2115234 0.5451634 0.8031325 0.9334468 0.9818109 0.9958714 0.9992053 [8] 0.9998681 0.9999809 0.9999976 0.9999997 |
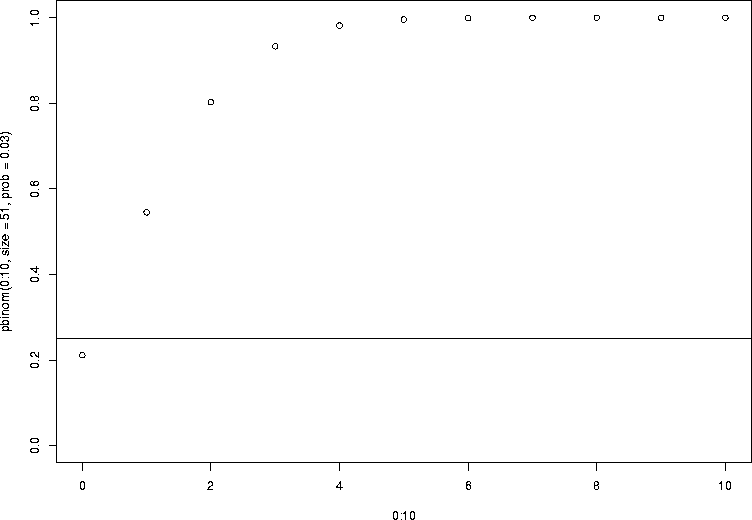
> qbinom(0.25,size=51,prob=0.03) [1] 1 |
So the purpose of the quantile function is to tell you where to draw the cutoffs to make sure you have a certain amount of the distribution on the left hand side. This is quite easy for a continuous random variable like the standard normal:
> qnorm(0.025) [1] -1.959964 > qnorm(c(0.005,0.01,0.025,0.05,0.1,0.5,0.9,0.95,0.975,0.99,0.995)) [1] -2.575829 -2.326348 -1.959964 -1.644854 -1.281552 0.000000 1.281552 [8] 1.644854 1.959964 2.326348 2.575829 |
R has built in quantile functions for all sorts of useful distributions that come up in statistics, eliminating the need for tables:
> qchisq(0.95,df=1) [1] 3.841459 > qt(0.025,df=10) [1] -2.228139 > qf(0.95,df1=3,df2=37) [1] 2.858796 |
Last time we spent some time talking about "exact" confidence intervals for the Binomial distribution. We observe a certain number X of successes in N independent idential bernoulli trials, and compute the observed frequency of success, X/N. And this value X/N estimates the true success probability. We found how to compute an interval estimate of this using the binomial distribution. Every time we do the experiment, we should get a different number of successes in principle, since this is a random variable, and therefore we should get a different confidence interval. The confidence interval is a random variable, and the idea is that it should have a certain chance of containing the true value. Let's simulate this process, as follows:
> cp.interval.lt <- function(xx,nn,alpha=0.05,low=1e-6,hi=1-1e-6) {
+ f1<-qf(alpha/2,df1=2*xx,df2=2*(nn-xx+1))
+ f2<-qf(1-alpha/2,df1=2*(xx+1),df2=2*(nn-xx))
+ b1 <- 1+(nn-xx+1)/(xx*f1)
+ b2 <- 1+(nn-xx)/((xx+1)*f2)
+ c(1/b1,1/b2)
+ }
> contains.val <- function(cis,a.value) {
+ (a.value > cis$lower) & (a.value < cis$upper)
+ }
> gen.intervals <- function(xxs,size,nvals=length(xxs)) {
+ lowers <- rep(NA,nvals)
+ uppers <- rep(NA,nvals)
+ for (ii in 1:nvals) {
+ intvl <- cp.interval.lt(xxs[ii],size,alpha=0.05)
+ lowers[ii] <- intvl[1]
+ uppers[ii] <- intvl[2]
+ }
+ lowers[is.nan(lowers)] <- -1
+ list(lowers=lowers,uppers=uppers)
+ }
> nn <- 20000
> nsims<-10000
> tru <- 0.5
> sum(contains.val(gen.intervals(rbinom(nsims,size=nn,prob=tru),nn), tru))
[1] 9535
|
> # continuing example from above > sum(contains.val( + gen.intervals(rbinom(nsims,size=nn,prob=tru),nn), tru + )) [1] 9805 There were 50 or more warnings (use warnings() to see the first 50) Rweb:> warnings() Warning messages: 1: NaNs produced in: qf(p, df1, df2, lower.tail, log.p) |